Galaxy
Galaxy is a platform for bioinformatic workflow management. Tools are a key component of Galaxy: they wrap a piece of software up with its dependencies so it can be simply deployed in any server without interfering with other tools. In that sense tools are share something with the conda package management system. But a Galaxy tool has other components specific to Galaxy: instructions for a gui input page, help contents and built-in test data. Complex analytical workflows can be constructed by combining tools using the Galaxy workflow manager capability (eg the image below). They can be stored and reused and shared with anyone with access to a galaxy server: reproducibility is built in to the analysis.
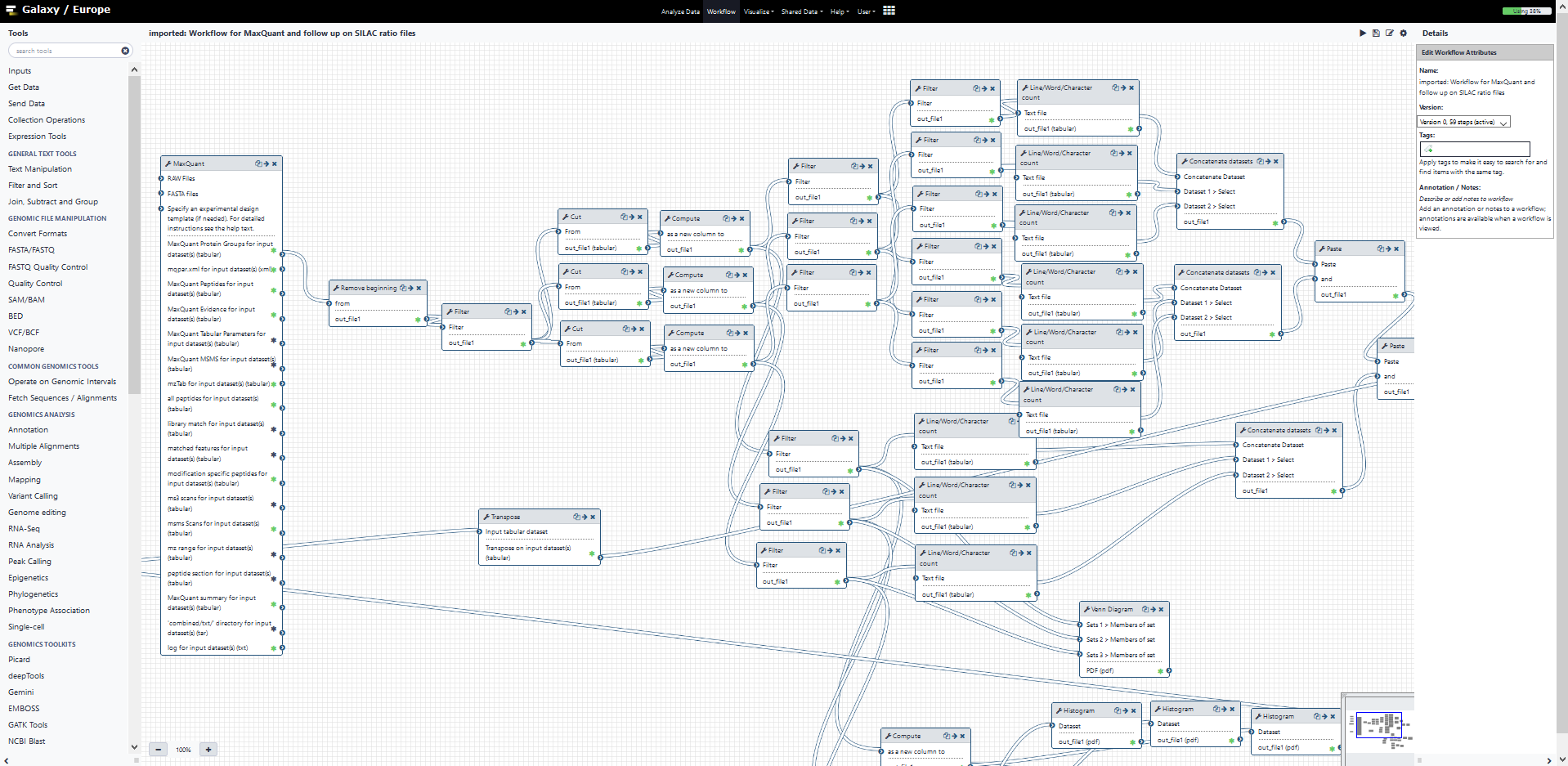
A simple tool example.
Tools have an .xml format reminiscent of the index.json file for conda packages. Below is skeleton of a tool file for a program called seqtk_seq.
<tool id="seqtk_seq" name="Convert to FASTA (seqtk)" version="0.1.0">
<requirements>
</requirements>
<command detect_errors="exit_code"><![CDATA[
TODO: Fill in command template.
]]></command>
<inputs>
</inputs>
<outputs>
</outputs>
<help><![CDATA[
TODO: Fill in help.
]]></help>
</tool>
